Evolutionary biophysics of cell polarity
Summary
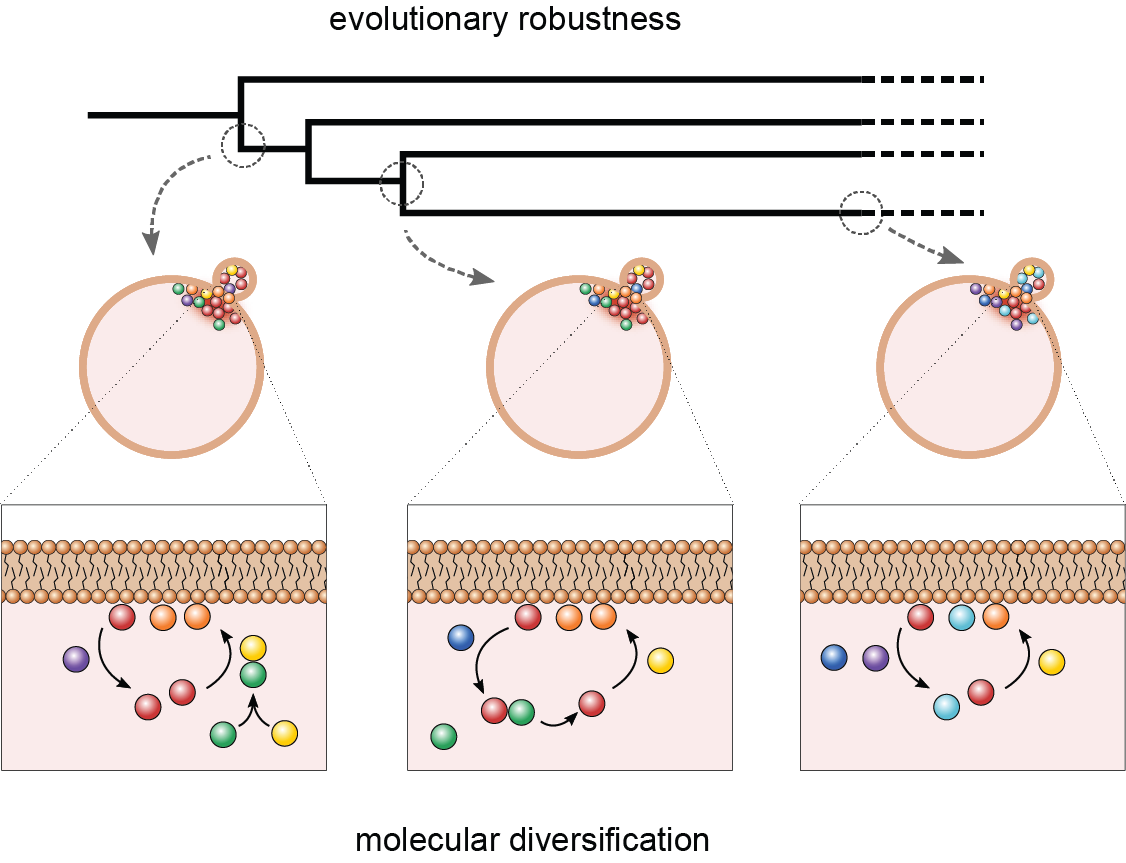
It is often assumed that essential proteins in one species are of similar importance across species. Surprisingly though, recent massive sequencing of non-model species showed that some proteins that are essential in model species are absent in related species, while the core function, e.g. cell polarity, metabolism or chromosome segregation is maintained over that branch of the tree of life. How is it possible that core cellular functions are conserved, while the responsible molecular networks dramatically diversify? We address this question using polarity establishment in Saccharomyces as a model system. Polarity establishment is essential for proliferation, directly affects cell fitness and thus needs to be maintained in every fungal species. However, in a comparative study of the polarity network between 298 fungal species, we found that the composition of the polarity network dramatically diversified over evolutionary time and even proteins essential in S. cerevisiae are regularly lost. These losses are typically explained by assuming that another protein replaces the function of the protein that is lost, while the rest of the network remains more-or-less unperturbed. Intriguingly though, it is becoming clear that diversification of core networks can also be driven by protein losses, which is difficult to understand from the perspective of one protein simply replacing another. How do these losses repair polarity? Work from our lab suggests that the polarity network collectively buffers protein loss through redundancies in pathways rather than through individual proteins. Our observations may make sense evolutionary: if selection acts on the establishment of cell polarity, rather than on specific molecules and if there is redundancy, than this opens up the possibility of changing the molecular composition, while maintaining the function. We are currently performing live cell microscopy and computational studies to understand this collective response.
Lab members
Marieke Glazenburg MSc.
Renske Voerman, BSc.
Frank van Opstal
Collaborators
Prof. dr. Erwin Frey, LMU, Germany
Dr. David Zwicker, Max Planck Institute Gottingen, Germany
Prof dr. Antonis Rokas, VanderBilt University, US
Prof. dr. Marko Kaksonen, University of Geneva, Switzerland
Dr Hylkje Geertsma, Imaging Physics, TU Delft
Papers
F Brauns, LM Iñigo de la Cruz, WKG Daalman, ..., L Laan# and E Frey# (2023)
Redundancy and the role of protein copy numbers in the cell polarization machinery of budding yeast
Nature communications 14 (1), 6504
# corresponding authors
MM Glazenburg, L Laan (2023)
Complexity and self-organization in the evolution of cell polarization
Journal of cell science 136 (2), jcs259639
David Zwicker and Liedewij Laan (2022)
Evolved interactions stabilize many coexisting phases in multicomponent liquids
Proc Natl Acad Sci, 119 (28) e2201250119 https://doi.org/10.1073/pnas.2201250119
Eveline T. Diepeveen, Valerie Pourquié, Thies Gehrmann, Thomas Abeel and Liedewij Laan (2018)
Patterns of conservation and diversification in the fungal polarization network.
Genome Biology and Evolution, evy121, https://doi.org/10.1093/gbe/evy121
Eveline T. Diepeveen, Leila Iñigo de la Cruz, and Liedewij Laan (2017)
Evolutionary dynamics in the fungal polarization network, a mechanistic perspective.
Biophysical Reviews 1-13 /doi/10.1007/s12551-017-0286-2
Liedewij Laan, John H. Koschwanez, Andrew W. Murray (2015)
Evolutionary recovery after crippling cell polarization reveals reproducible trajectories.
eLife, 10.7554/eLife.09638